Clinical practice is beginning to be transformed by a new era of ‘omics’ – genomics, epigenomics, pharmacogenomics, transcriptomics, proteomics, metabolomics, lipidomics, and so on – based on large patient biodata sets. The following review describes how these diverse molecular technologies are being applied to the clinical practice and research of cancer medicine.
Each biomolecular class – RNA, DNA (both of which are nucleic acids), protein or lipid – has become the focus of its own field of academic study or ‘omic’, a suffix derived from the Greek word for ‘body’ (in this context denoting a body of knowledge). The disease-related information obtainable from these different molecular classes is complementary,1 raising the hope that future syntheses of this broad data spectrum will provide better therapeutic predictivity for molecular targets of anticancer therapies.2,3 What we now designate biomarkers4 may prove to be the first steps towards this goal.5 Examples of molecular testing currently pertinent to clinical practice or research in oncology are shown in Table 1. As matters stand, however, the quantity of information available from these data sources far exceeds our ability to exploit it therapeutically, partly reflecting the relative paucity of treatment options available for such customisation.6
Table 1. Examples of molecular testing pertinent to clinical practice or research in oncology
|
Molecular species
|
Alteration of interest
|
Abnormality in cancer
|
Example
|
|---|
|
RNA
|
Change in gene expression
|
Induction
|
PI3K, AKT
|
|
Repression
|
CDKN1A
|
|
Gene expression profiling
|
Distinct co-expression patterns
|
Luminal A versus triple-negative breast cancer
|
|
Gene mutation
|
Missense
|
TP53. KRAS
|
|
Nonsense
|
BRCA2
|
|
DNA
|
Gene copy number variation
|
Amplification
|
HER2, MYC
|
|
Loss of heterozygosity
|
APC, PTEN
|
|
Karyotype
|
Chromosomal translocation
|
9:22
|
Chronic myeloid leukaemia
|
|
|
Chromosomal deletion
|
3p21
|
Small cell lung cancer
|
|
Protein
|
Change in protein expression
|
Upregulation
|
ERK
|
|
|
Downregulation
|
pRb
|
|
Lipid
|
Change in (extra)cellular lipids
|
Increase
|
Phosphatidylcholine
|
|
|
Reduction
|
Phosphatidylethanolamine
|
Genomics
The term ‘genomics’ denotes the study of the structure and function of DNA. The somatic genome of a human cell or tissue accrues damage because of ageing, whereas the germline (familial) genome is passed down through successive generations. Germline disease genomics are relevant to familial cancer syndromes, such as BRCA-mutant breast/ovarian cancer, whereas, somatic genomics can help elucidate the pathogenesis of sporadic cancers. Germline/familial gene defects are rarer than the same defects in somatic tumours; for example, BRCA1/2 gene mutations are very often present in breast cancers of patients from families without the BRCA mutation. Similarly, absence of mismatch-repair enzyme expression is frequently evident in sporadic colorectal cancers from patients who do not hail from Lynch syndrome families.
Single-gene testing
Single-gene disorders can be identified using sequencing limited to coding gene (ie exomic) mutations.7,8 In the case of cancer, exomics has focused on two broad functionally defined gene categories: cancer-causing ‘oncogenes’ that drive cell growth and anti-cancer tumour suppressor genes.9 The latter include DNA repair genes needed for maintaining genetic stability – defects in such genes may predispose to mutation-prone ‘microsatellite-unstable’ colorectal cancers that more often respond to immunotherapy.10
Multi-gene testing
The advent of next-generation sequencing (NGS) has made plausible the ‘thousand-dollar genome’ for either germline studies or somatic disease, such as cancer. Multi-gene tumour mutation screens are already available. Such assays assess up to 500 relevant genes per panel, including cancer ‘drivers’ (oncogenes; eg KRAS, BRAF) and ‘suppressors’ (eg TP53, MLH1). Although these tests are of high research interest, there can be few guarantees for today’s patients as to any proven or cost-effective improvements of disease outcomes.11
The Cancer Genome Atlas
In 2005, the National Cancer Institute in the US received funds to begin The Cancer Genome Atlas (TCGA) project, which involves sequencing all common cancer-related genomic aberrations. For research purposes, TCGA genome sequencing data are now accessible for common human tumours, such as glioblastoma multiforme, breast cancer, colorectal and stomach cancer, ovarian and uterine cancer, melanoma, myeloid leukaemia, lung and head/neck cancers, and renal and bladder cancers. Computational recognition of such tumour-specific genomic ‘signatures’ should in future help doctors to assign the most appropriate treatments to metastatic disease of poor differentiation and/or unknown primary origin.12
Pharmacogenomics
Pharmacogenomics is the study of germline genomic variants that give rise to unusual drug sensitivity (benefit) or resistance/toxicity. For example, patients with CYP2D6 duplications may be hypersensitive to codeine toxicity. Once personalised genome information is routinely available, such information may be programmed into electronic medical record (EMR) systems to provide interactional warnings. Repositioning of available drugs for new disease-related uses could also prove to be a value-adding application of pharmacogenomics.13
Single-cell genomics
Tumour biopsies may not be essential for cancer diagnosis in the future. New technologies can identify and analyse either circulating cell-free DNA from tumours or circulating tumour cells.14 These non-invasive methods offer the prospect of ‘liquid biopsies’ in preference to tissue sampling, although remaining an area of research interest for now. Technically related to these single-cell advances is the use of cancer-specific ligands for diagnostic or therapeutic purposes. This has long been exploited for the treatment of metastatic differentiated thyroid cancer using radio-iodine and, more recently, for metastatic low-grade neuroendocrine tumours (using octreotide-linked radio-lutate).15 Positron emission tomography (PET) scans using radiolabelled antibodies could likewise deliver non-invasive diagnoses, while also providing organ-specific immunoimaging of metastatic protein expression.16
Epigenomics
The reversible (ie non-mutational or epigenetic) modification of human DNA, genes or chromatin structure by chemical alterations, such as methylation or acetylation, can affect gene expression in a heritable manner. The most studied field of epigenomics has been the clustered dinucleotides known as CpG islands within cancer-related gene promoters.17 Methylation of such CpG islands leads to loss of transcriptional initiation and hence expression18 of wild-type tumour suppressors. However, only a few areas of epigenomics are clinically relevant at present. The DNA methyltransferase inhibitor azacitidine is used to treat myelodysplastic syndromes, although the extent to which its efficacy reflects epigenetic versus cytotoxic effects remains unclear. Promoter methylation of the tumour suppressive DNA repair enzyme MGMT (O6-methylguanine DNA methyltransferase) appears to be predictive for therapeutic efficacy of the cytotoxic drug temozolomide in the treatment of glioblastomas.19
Transcriptomics
Gene expression studies (transcriptomics) are one of the oldest omics, as it is technically simple to isolate cellular RNA. Given that only a subset of genes is expressed in any differentiated tissue and the transcriptome is smaller than the exome, it may be more informative from a functional (as distinct from mutational) perspective. The original way to characterise transcriptomes was to use ‘gene chips’ or cDNA microarrays. This approach distinguishes clinically informative gene expression subsets in breast cancer (eg HER2+ versus triple-negative breast cancer) and colorectal cancer. Today’s researchers prefer to use another expression profiling approach termed RNA-Seq (‘whole transcriptome shotgun sequencing’), which offers far greater informational specificity than microarray approaches. Relevant to this technological advance, it has recently become clear that transcriptomes include functionally critical, long, non-coding (untranslated; interfering) RNAs arising from intergenic (non-gene) sequences, and variants related to gene splicing and fusion events.20
Several commercial gene expression panels are now being marketed to oncologists who treat conditions such as node‑positive primary breast cancer.21 Some of these expression profiles calculate the patient’s relapse risk on the basis of over-expression or under-expression of particular genes involved in cancer growth or regulation. The decision-making value of such panels often focuses on whether patients should receive adjuvant chemotherapy.
Metabolomics and lipidomics
Patterns of plasma lipid abundance are referred to as the lipidome and have long been known to affect rates of cardiovascular disease.22,23 More recently, genetic and diet-related changes in the lipidome have also been implicated in the pathogenesis and progression of neoplastic disorders,24 such as breast cancer.25 The involvement of either plasma or cellular lipids in metabolic regulation represents one key part of the larger field of metabolomics, which is starting to include newer subsets such as nutrigenomics.26
Proteomics
Protein-based histopathological assays, such as immunohistochemistry, have been the traditional basis of laboratory-based tumour characterisation, but are limited to those disease-related antigens for which validated antibodies already exist. Developing a more discovery-based approach to the full set of proteins expressed by cancer cells27,28 has required harnessing mass spectrometry,29 which is a cumbersome and labour-intensive platform that has not lent itself to routine clinical use.30 Since protein function is often altered by reversible post-translational modifications, such as phosphorylation, phosphoproteomic analyses using antibodies specific for different phosphorylation state31 may provide future insights into signalling pathways of therapeutic relevance.32,33
Phenomics
A newcomer to the omics universe is phenomics – the study of phenotypes, and how traits are affected by genetic and environmental change.34 Phenomics may thus be viewed as a mirror image of functional genomics, originating as it does, not from a gene of interest, but from a target behaviour. The challenge for the phenomic field is to create high-throughput platforms that collect quality data in the background, such as clinical details from EMR systems using text-mining,35,36 and thus permit correlation with omic-derived or external variables (Figure 1).37 Phenomics may thus be central to the future of clinical decision support systems.
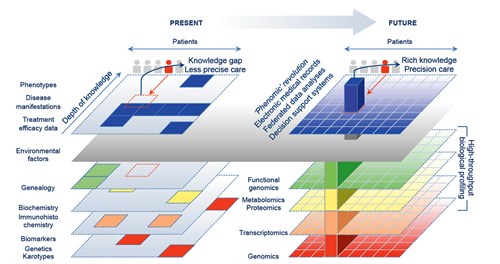
Figure 1. From phenotypes to phenomics – how clinical information is changing
Left: Current low-throughput methods result in biomedical knowledge gaps, with clinical decisions based on incomplete data, and treatments tailored through crude or empirical criteria. Right: The ‘omics revolution’ promises a knowledge-enriched way to tailor treatments through integrating rich informatic profiles with personalised matching of molecular patient data to help fine-tune precision healthcare
Conclusion
Recent technological advances have enabled us to unlock the secrets of the cancer genome. Although clinical applications are currently limited, the next decade is likely to see significant progress based on better understanding of this uniquely personalised information repository.
Authors
Richard J Epstein MBBS, MD, MBA, PhD, FRCP, FRACP, Senior Consultant, Department of Oncology, St Vincent’s Hospital, Darlinghurst, NSW; Conjoint Professor, UNSW Clinical School, Kensington, NSW; Visiting Scientist, Garvan Institute for Medical Research, The Kinghorn Cancer Centre, Darlinghurst, NSW. Richard.Epstein@svha.org.au
Frank P Lin MBChB, PhD, Senior Research Officer, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Darlinghurst, NSW; Oncology Registrar, St Vincent’s Hospital, The Kinghorn Cancer Centre, Darlinghurst, NSW; Conjoint Lecturer, UNSW, Kensington, NSW
Competing interests: None
Provenance and peer review: Commissioned, externally peer reviewed.
Acknowledgements
We thank St Vincent’s Hospital and the Garvan Institute for their kind support.