Commercial DETs exist, but, like EHRs, they are largely proprietary ‘black-box’ solutions with intellectual property protection preventing adequate assessment of any design or execution errors or quality of data extracted. Effective assessment and management of data quality (DQ) requires analysis of the whole data cycle: from collection through extraction, cleansing, storage, management, dissemination, presentation and curation.10 Data quality management (DQM) processes and information governance (IG) structures are needed to ensure that data routinely captured within clinical practice is complete, correct, consistent11 and, ultimately, is fit for purpose.12
We examined whether different DETs achieved consistent results. Diabetes was used as the exemplar because it has a known prevalence, is clinically important and should be consistently extracted from EHRs as the diagnosis is based on numeric data13 and most anti-diabetic drugs and pathology tests are diabetes-specific. United Kingdom researchers have set out the sensitivity and specificity of surrogate markers of diabetes14 and differentiated between people with poor DQ within their EHRs, subdividing them into those who have errors in coding, classification or diagnosis of their diabetes. Around 40% of people with one or more of these errors have underlying clinically significant issues15 and those not included in computerised patient registers seem to receive worse care.16
The University of New South Wales (UNSW) electronic Practice-Based Research Network (ePBRN)7 compared ‘DET1’, its in-house data extraction and linkage tool,17,18 to two other DETs. We tested the hypothesis that the counts of the diabetes cases identified and their demographic and clinical data extracted from a general practice EHR will be the same for DET1 and two other proprietary DETs.
Methods
Two different EHRs (EHR1 and EHR2), were used to compare DET1, which extracts from both EHRs, with:
- DET2, which extracts from EHR1 and EHR2
- DET3, which only extracts from EHR1.
EHR1 uses a proprietary coding (terminology) system while EHR2 uses the International Classification of Primary Care version 2 (ICPC2).19 Both EHRs allow free text entry if the codes are not available. Electronic health records are typically relational databases with a number of linked data tables, including: ‘History’, ‘Past History’, ‘Diagnosis’, ‘Medication’, ‘Pathology’, ‘Measures’ and so on. The data models vary as there are no prescribed standards. In EHR1, the ‘Diagnosis Table’ and ‘History Table’ capture diagnostic terms from ‘reason for visit’, ‘prescription’ and ‘procedure’ data entry windows.
The location of key-coded data varies from EHR to EHR, and clinicians may be unclear as to the consequences of their recording choices, or even of what the default settings might do. The following examples set out how apparently innocent choices about where data are stored or saved might affect whether a case, risk factor or treatment might be identified by a subsequent search:
- The ‘reason for contact’ that can include a problem or diagnosis is saved in the ‘Diagnosis Table’. If ‘save in past history’ is ticked, the default option in EHR1, it is also saved in the ‘History Table’. In EHR2, the ICPC2 codes are captured in the ‘Problem Table’ and glycosylated haemoglobin (HbA1c) tests in the ‘Pathology Table’.
- DETs often use different tables or a number of tables as the data source, eg. DET2 and DET3 identified diabetes cases from the ‘condition’ attribute in the ‘History Table’ of EHR1, while DET1 used the ‘reason for contact’ attribute in the ‘Diagnosis Table’. DET2 and DET3 also ‘transformed’ the extracted data, using a proprietary tool.
The extractions were run one after the other for each EHR (Figure 1). DET1 used an extensible mark-up language (XML)-specified query to extract data. For DET2, relevant terms were entered into the search window to identify patients flagged as active in the EHR (EHR-active) and pre-programmed reports generated. For DET3 the pre-programmed reports were executed. All extracted data were sent to a data repository where they were analysed using Statistics Package for the Social Sciences (SPSS) software (version 20.0; IBM SPSS, Armonk, NY, USA). DET1 used a comprehensive list of approximately 300 structured and free text terms used to describe diabetes in both EHRs; DET2 and DET3 used the relevant coded terms available for EHR1, but the documentation was not explicit about the actual terms used. For EHR2, DET1 and DET2 used ICPC219 codes for diabetes and diabetes-related conditions (T89, T90, T31, T45, T68, T99, N94, F83 and the relevant 6-digit extensions).
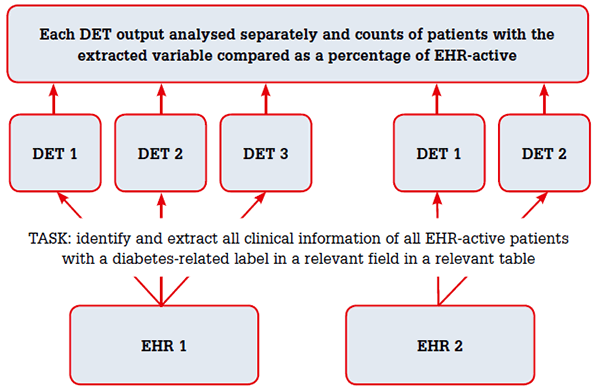
Figure 1. Methodology for comparison of DETs output
The causes of the variations were categorised into clinical practice, EHR, DET and data quality factors. This built on an existing taxonomy of information technology (IT)-related data extraction errors,20 which categorised errors in extracted data into: (1) extraction method and process, (2) translation layer, (3) shape and complexity of original schema, (4) communication and system (software) faults, (5) hardware and infrastructure, and (6) generic or human errors.
Results
The three DETs delivered different counts of ‘EHR-active’ patients, diabetics and quality (completeness) of diabetes data (Table 1). Differences ranged from 0.1% to 10%, some statistically significant, for active patients; diabetics identified by condition; HbA1c; diabetes medication; and risk factors such as smoking status, body mass index (BMI) and blood pressure (BP).
EHR-active patients
The number of designations for EHR-active patients differed significantly, with up to 10% difference between the DETs. Possible explanations include incomplete or untimely flagging of EHR-active status by staff/general practitioners (GPs); inconsistent use of patient status codes (such as death, transferred or deleted) and search queries (such as how the DET/EHR handles the options for patient status, empty data fields or the ‘next-of-kin’ field); incompatibility of the DET/EHR data models; corrupted interactions of the DET/EHR software routines; and data corruption for technical and functional reasons.
Identifying diabetes by specific database tables
The number of diabetics identified by a diagnostic label varied by DET and source table used. (a) EHR1: DET1 identified more diabetics than the other DETs from the ‘History Table’ but fewer from the ‘Diagnosis Table’. Because DET2 and DET3 used the same coded terms, they extracted nearly identical numbers from the ‘History Table’. DET1 extracted more medications than DET3 from EHR1, largely due to differences in terms/codes used for the diabetes medication queries. DET1 used the therapeutic class (THERCLASS) codes of EHR1, whereas DET3 identified medications in EHR1 by what they called ‘mappings’ to the THERCLASS but did not provide details. (b) EHR2: DET1 extracted more diabetics (using ICPC2 codes) than DET2 from the ‘Problem Table’ and HbA1c tests from the ‘Pathology Table’.
Identifying diabetes using multiple tables
By combining the information from multiple tables (‘Diagnosis’, ‘Pathology’, ‘Prescriptions’ and ‘Measures’) and eliminating duplicates, DET1 identified more diabetics than when using a single table (Table 1). DET2 and DET3 identified patients only from the ‘History’ table.
Table 1. Variations in a sample of data extracted from two EHRs by three DETs under the same experimental conditions
| Data extracted from specific tables in EHR: | DET1 | DET2 | DET3 |
|---|
| Patients n (%) | 95% Confidence intervals | (CI)Patients n (%) | 95% CI | Patients n (%) | 95% CI |
|---|
| 1. EHR1 |
| All EHR-active patients |
21 793 |
N.A. |
24,145 |
N.A. |
24,180 |
N.A. |
| Patients with diabetes-related diagnostic labels |
558 (2.9) |
2.69–3.13 |
598 (2.5) |
2.29–2.68 |
599 (2.5) |
2.29–2.68 |
| Patients with HbA1c tests |
296 (1.4) |
1.21–1.52 |
253 (1.0) |
0.90–1.80 |
253 (1.0) |
0.93–1.18 |
| Patients with diabetes-related prescriptions |
642 (3.0) |
2.73–3.18 |
Did not extract routinely |
485 (2.0) |
1.84–2.19 |
| Patients with diabetes-related diagnostic labels, tests or prescriptions |
833 (3.8) |
3.60–4.10 |
DET2 and DET3 do not routinely extract from multiple tables |
| 2. EHR2 |
| All EHR-active patients |
25 770 |
N.A. |
25,770 |
N.A. |
DET3 does not
extract from EHR2 |
| Patients with diabetes-related diagnostic labels |
367 (1.4) |
1.29–1.58 |
234 (0.9) |
0.80–1.03 |
Demographics
A similar random variation pattern was found, which may reflect differences in the diabetics identified, data-entry options for gender/date of birth or how the EHR handles data storage and exchange between EHRs and linked billing systems, usually separate proprietary software. Incomplete or incorrect data entry may be an issue as one patient had an EHR default date of ‘01 Jan 1800’ in the date-of-birth field.
Risk factors
All three DETs extracted different numbers of diabetics with a recorded BMI or BP from EHR1. When expressed as a proportion of the numbers of diabetics the DET identified, DET1 extracted a lower proportion across all the risk factors.
Discussion
The DET/EHR data models were proprietary and not transparent. However, we gained sufficient insight into the DET/EHR from available documentation, iterative use and discussions with vendor technical support and members of the ePBRN. Generally, DETs extract, store, manipulate and report on a snapshot of coded data in the EHR. The data model, data and metadata are not tailored or updated systematically or validated independently or in association with the EHR. As such, there are often mismatches between the DET and EHR data models. The ePBRN experience with repeated data extractions suggests that EHRs are often not consistent in how they store codes or data over time.
The sociotechnical conceptual framework describes how users and technology undergo a process of mutual transformation. Flagging variations in extracted data in terms of technical and system design factors, differing practices in documentation, workflow and related factors may improve system design and more consistent recording of data.21 This study highlights the variation between each DET/EHR combination, and sets a potential agenda for improving our ability to monitor quality.
A limitation of this study is that it reported crude extract numbers. None provided a ‘gold standard’ extraction that matches the expected 6.6% prevalence of diabetes in the study regions, as reported in the South Western Sydney Local Health District 2012 Annual Report. While we do not provide adjusted prevalence figures, it is plausible that some DETs are missing large numbers of relevant patients.
We conclude that the DET/EHR combinations did not extract similar counts of diabetics and indicators of diabetes care. This renders current DETs ineffective as tools for measuring the quality of care in a way that might be compared between systems. When we add the lack of transparency for proprietary reasons and a lack of technical and professional standards and safety regulations for medical software, this situation is unable to ensure that practice is safe, or able to support clinical governance.
Organisations promoting eHealth must be accountable and transparent22 and their software products subject to appropriate and independent accreditation and regular review, including monitoring for critical incidents associated with their use.
Implications for general practice
- Data extracted from EHRs may be unreliable.
- EHRs and extraction tools must support independent testing with standardised queries.
- The proprietary model for software quality control is not in our best interests.
- Appropriate information governance processes and structures must be established.
Competing interests: Siaw-Teng Liaw has intellectual property in DET1, but was not involved in the conduct of the experiment reported in this study. This research was funded in part by a seeding grant from the School of Public Health & Community Medicine, University of New South Wales.
Ethics approval: This study has approval from the UNSW Human Research Ethics Committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
Acknowledgements
The authors wish to thank Dr Douglas Boyle, Mr Ian Peters and Ms Yin Huynh for advice on the DETs, and Dr Blanca Gallego-Luxan and Prof Simon Jones for comments on drafts.