More than 90% of cases of acute diarrhoea are caused by infectious agents1 acquired by faecal-oral transmission via direct personal contact or ingestion of contaminated food or water. It is important to have an understanding of the range of pathogens that may cause infectious diarrhoeal syndromes (Table 1–3).
Table 1. Clinical infectious diarrhoeal syndromes
| Syndrome | Clinical features | Epidemiology | Typical pathogens |
|---|
| Watery diarrhoea |
Loose, watery stools, no blood |
Most common presentation |
- Campylobacter spp.
- Salmonella spp.
- Shigella spp.
- Vibrio spp.
- Yersinia spp.
- Plesiomonas spp.
- Aeromonas spp.
- Most viruses and parasites
|
| Bloody diarrhoea |
Bloody stools, sometimes mucous or pus
Often abdominal pain, fever, tenesmus |
Less common |
- Shigella spp.
- Shiga-toxin producing Escherichia coli (STEC)
- Salmonella spp.
- Campylobacter spp.
|
Table 2. Infectious diarrhoeal syndromes in particular epidemiological settings
| Travellers’ diarrhoea |
Loose, watery stools sometimes bloody |
Visitors to developing tropical/semi-tropical countries |
- Most commonly enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC)
- Campylobacter spp.8
- Salmonella spp.
- Viruses and parasites
|
| Nosocomial diarrhoea |
Spectrum of disease; mild diarrhoea, fulminant colitis, toxic megacolon
Healthcare and antibiotic associated diarrhoea9 |
Risk factors:
- hospitalisation
- elderly
- antibiotic therapy3
Community acquired disease (increasingly common2) |
- Toxigenic Clostridium difficile
- Norovirus
|
| Diarrhoea in immunocompromised patients |
Variable presentation
Careful investigation required |
- HIV patients
- Solid organ/bone marrow transplant recipients
- Cancer patients undergoing chemotherapy
|
- Salmonella spp.
- Shigella spp.
- Campylobacter spp.
- Atypical mycobacteria10
- Viruses and parasites10
|
Table 3. Bacterial pathogens that cause acute diarrhoea
| Pathogen | Major modes of transmission | Clinical features | Epidemiological features |
|---|
| Campylobacter jejuni |
Food borne |
- Watery diarrhoea
- Can cause fever and bloody diarrhoea
|
- Very common pathogen
- Associated with undercooked poultry
- Common cause of travellers’ diarrhoea
|
| Nontyphoidal Salmonella spp. |
Food borne |
|
- Common pathogen
- Associated with undercooked poultry, eggs, other meat products
|
| Shigella spp. |
Person-to-person
Food and water borne |
- Often severe diarrhoea; bloody stools, fever, abdominal pain
|
- Human reservoir
- Low infectious inoculum
- Secondary cases common within households
- Most severe disease:
S. dysenteriae – mainly found in developing countries
|
| Yersinia enterocolitica |
Food borne |
- Usually watery diarrhoea
- Can produce fever, bloody diarrhoea
- Can mimic acute appendicitis
|
- Animal reservoir (especially pigs)
- Associated with pork products
|
| Shiga-toxin producing E. coli (STEC), includes E. coli 0157 |
Food borne |
- Watery diarrhoea – can progress to bloody diarrhoea
- Important cause of haemolytic uraemic syndrome
- Children and the elderly
|
- Reservoir in cattle
- Associated with undercooked beef (especially ground beef)
|
| Vibrio spp. |
Food borne (especially shellfish) |
- Watery diarrhoea
- Can produce bloody stools
|
- Associated with shellfish, other seafood (particularly prevalent in warmer months)
|
| Plesiomonas spp. |
Food and water borne |
- Watery diarrhoea
- Can produce bloody stools
- Severe disease seen with liver disease and malignancy
|
- Associated with overseas travel, consumption of shellfish and other seafoods
|
| Aeromonas spp. |
Food and water borne |
- Acute watery diarrhoea, bloody diarrhoea and chronic diarrhoea
- Severe disease seen with liver disease and malignancy
|
- Aquatic environmental reservoir
|
Salmonella enterica serovars Typhi and
Paratyphi |
Food and water borne |
- Systemic toxic effects, abdominal symptoms, fever with or without diarrhoea
- Causes bacteraemia
|
- Mainly travellers to developing countries
- Contaminated food and water
- Human reservoir
|
| Clostridium difficile |
Bacteria and spores in the hospital environment (eg. the hands of staff, fomites including benchtops and surfaces) |
- Spectrum of disease; usually watery diarrhoea, can be bloody
- Toxic megacolon, perforation and death can occur
|
- Most common cause of healthcare and antibiotic associated diarrhoea9
- Community acquired disease – increasingly recognised2
|
Indications for testing
Most cases of acute diarrhoea are mild and self limiting and no investigation or treatment is necessary. However, some patients should be investigated regardless of the severity of disease: returned travellers, patients in whom diarrhoea has persisted for more than 4–5 days, patients with bloody stools, immunocompromised patients, and in cases where there is suspicion of an outbreak of enteric disease. Admission to hospital is usually only required in cases of significant dehydration, marked toxaemia, persistent vomiting or severe abdominal pain.
In patients with severe symptoms, empirical antibiotic therapy may be appropriate pending the results of laboratory investigations.1
Logical positioning of stool cultures relative to other related investigations
A thorough clinical history and examination is essential before requesting stool culture. Nonbacterial causes of diarrhoea (eg. viral or parasitic) should be considered in the differential diagnosis. Viruses are more common in children (particularly rotavirus and adenovirus) and are usually self limiting. Norovirus is also an important cause of community acquired diarrhoea. Nucleic acid amplification testing or antigen detection assays are available for detection of viruses in the faeces, but are rarely indicated in the outpatient setting. Testing for parasites (eg. Giardia, Entamoeba) should be prompted by specific epidemiological risk factors (eg. travel, immunocompromised host). For parasites, diagnostic options include stool microscopy for ova, cysts and parasites and antigen detection assays.
Clostridium difficile is an increasingly recognised cause of community acquired diarrhoea.2 Risk factors include (but are not restricted to) advanced age, recent hospitalisation and recent antibiotic use.3 Where C. difficile associated disease is suspected, a stool specimen should be submitted for culture and/or antigen screening.
‘Food poisoning’ occurs as a result of ingestion of pre-formed bacterial toxins present in food. It is self limiting and specific diagnostic testing is generally not required.
What should I tell my patient about the test?
Specimen collection: stool specimens submitted for testing should be loose or unformed, as many laboratories will not process formed stools. Stool specimens are preferred over rectal swabs. The patient should pass the stool into a clean, dry pan or a container mounted on the toilet. 5 mL of diarrhoeal stool, or 1–2 cubic centimetres of faeces, should then be transferred into a sterile container with a screw-top lid. Care should be taken to avoid any contamination with urine or toilet paper, and the specimen should be transported as quickly as possible to the laboratory. The risk of faecal-oral transmission of pathogens necessitates careful hand hygiene after the collection and handling of faecal specimens. In situations where delays in transport are anticipated or commonly encountered (eg. in remote areas) specimens should be kept at 4°C in a dedicated refrigerator. Stool specimens should never be frozen. Specimen jars containing a transport medium such as Cary-Blair can also be used to help maintain the viability of pathogens.
Some laboratories may reject specimens, if received more than 2 hours after collection, without utilising transport medium. Delays in processing of specimens can affect the recovery of some bacteria, such as Shigella species.
Timing of the test
Faecal specimens should be collected as early as possible in the course of the illness as pathogens decrease in number with time. If more than one specimen is submitted, these should be collected on separate days. The laboratory usually reports culture results within 3 days of receipt of the specimen.
Medicare eligibility and/or costs for the patient
Microscopy and culture of faeces, pathogen identification and susceptibility testing (a single examination in a 7 day period) is fully covered for eligible patients by the Medicare Benefits Schedule (MBS).
How does the test work?
On receipt of the specimen in the laboratory, the gross appearance of the specimen is inspected for consistency and for the presence of blood or mucous.
Microscopy is then performed to examine for erythrocytes and leucocytes and to screen for ova, cysts and parasites. A full examination for ova, cysts and parasites should be specifically requested if a parasitic infection is suspected, as the laboratory must process these specimens using specific methods.
Some laboratories may also perform a lactoferrin test for the detection of faecal leucocytes. Lactoferrin is an iron binding glycoprotein found in leucocyte granules, which is used as a marker for the presence of leucocytes in stool.
Culture is performed by inoculating faecal material onto a combination of different agar culture plates. These comprise selective and differential media that are used for the isolation and preliminary identification of specific organisms. Suspected pathogens are then formally identified to species level using a range of manual and automated methods.
Laboratories routinely culture for Salmonella, Shigella and Campylobacter species.4 Testing for other pathogens, such as Yersinia enterocolitica or Shiga-toxin producing E. coli and C. difficile requires special laboratory techniques.
Antibiotic susceptibility testing is usually only performed and reported for organisms that tend to produce more severe disease and more commonly require antibiotic therapy, such as Shigella and serovars Typhi and Paratyphi of Salmonella enterica. Salmonella isolates are referred to a reference laboratory for serotyping for public health purposes.
What do the results mean?
Stool culture detects the presence of any potential pathogens in the specimen.
What won’t it tell you?
One specimen is usually sufficient for the detection of most bacterial pathogens. However, submitting a second specimen has been shown to increase the overall sensitivity of the test by 20%.5 Submission of two consecutive specimens has been reported as sufficient to detect 99% of bacterial agents.6 The test cannot differentiate between colonisation (ie. asymptomatic carriage) and disease. Asymptomatic carriage does occur with some bacteria, for example C. difficile and Salmonella species. Thus, as with any test, the result must be interpreted in the clinical context. Antibiotic susceptibility results are not always reported as most pathogens (eg. Campylobacter) have relatively predictable susceptibility profiles. This also serves to discourage inappropriate antibiotic treatment.
What are the common next steps if the test is positive?
Most episodes of bacterial diarrhoea are self limiting and management is mainly supportive. Antibiotic treatment of bacterial diarrhoea is an area of considerable debate and is beyond the scope of this article. However, a positive stool culture should not necessarily prompt antibiotic therapy: excellent guidelines for the diagnosis and management of infectious diarrhoea are available through the Infectious Diseases Society of America website (www.idsociety.org). Australian guidelines for the management of bacterial diarrhoea are available in Therapeutic Guidelines, Gastrointestinal7 (www.tg.org.au).
In many jurisdictions, certain diarrhoeagenic enteric pathogens are notifiable to the public health authorities. It is the responsibility of the requesting practitioner to notify these authorities on receipt of the laboratory result. The need to notify an infection should be specified at the end of the laboratory report.
What if the result is negative?
If stool cultures are negative and diarrhoea persists, consideration should be given to clinical re-evaluation and/or additional investigations for nonbacterial causes (eg. parasites, viruses) and noninfective causes of diarrhoea (eg. inflammatory bowel disease or adverse effects of medication). This may include stool examination for ova, cysts and parasites, antigen detection tests, nucleic acid amplification tests, blood tests (eg. full blood count including peripheral blood eosinophilia, parasite serology, serum chemistry) and endoscopy. In cases where an infectious cause is suspected but routine laboratory testing fails to identify a pathogen, discussion with a clinical microbiologist or an infectious diseases physician may be appropriate, particularly when considering further testing or empirical treatment.
Special features of the test
Thorough clinical details are essential to help the laboratory perform the appropriate type of stool cultures. It is important to ensure that these are written legibly on the request form. Appropriate clinical information includes the age of the patient, whether the patient is immunocompromised, history, location and timing of foreign travel, recent consumption of shellfish or seafood, presence of blood in stools, fever or abdominal pain and recent antibiotic therapy.
Reporting of results
An illustrative laboratory report is shown in
Figure 1.
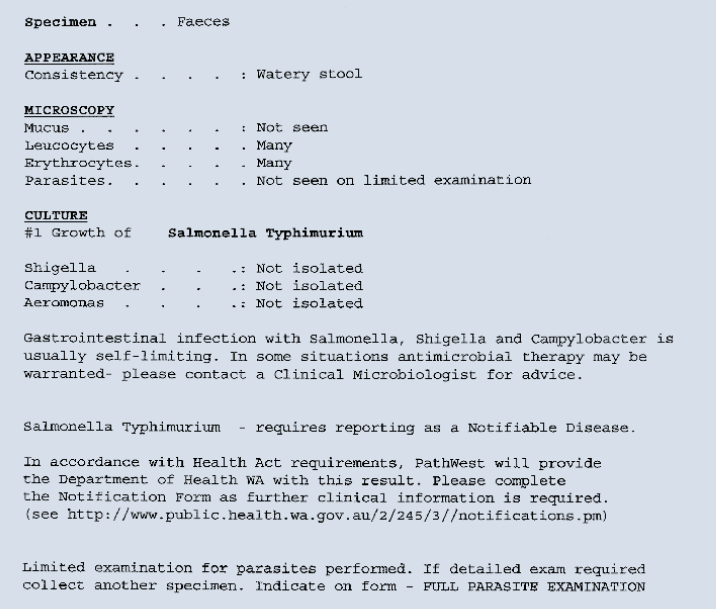
Figure 1. Illustrative laboratory report
Summary
Stool culture plays an important role in the investigation of the patient with suspected infectious diarrhoea. It is important to have an understanding of the most common pathogens and to appreciate that provision of adequate clinical information to the laboratory is critical to ensure that the appropriate stool testing is performed.
Declaration: Christopher Heath has been on the Antifungal Advisory Board of Gilead Sciences Inc (Aust), Merck Sharp & Dohme (Aust), Pfizer and Schering Plough and has received payment for expenses for attending IDSA and ICCAC meetings from both Merck and Pfizer.
Acknowledgements
The authors thank Mr Brian MacKenzie, Enteric Laboratory, PathWest Laboratory Medicine, Sir Charles Gairdner Hospital, Nedlands, Western Australia.